A review of the status of genomic applications and perspectives for fisheries and aquaculture

The development of sequencing technologies [the process of determining the nucleic acid sequence (the order of nucleotides, the basic building blocks, in the DNA of an organism) and the advances of genomics [interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes; a genome is an organism’s complete set of DNA, including all of its genes] is instrumental in addressing some of these challenges and can benefit sustainable fisheries and aquaculture.
With the development and advancement of massive parallel sequencing [any of many high-throughput approaches to DNA sequencing; known as next-generation or second-generation sequencing] technologies originating around 2005, more than 200 fish genomes have been sequenced and made available in public repositories. For example, as of late 2019, some 270 assembled fish genomes were available at the U.S. National Center for Biotechnology Information NCBI Genome.
These and various other genomic resources promote not only basic sciences such as comparative genomics, evolution, and systematics but also applied practices in aquaculture and fisheries, and all known species of fish (more than 34,000 species recorded in FishBase) will soon have their genomes sequenced. The subsequent challenges are how to make use of these genomic data and transform genomic knowledge into fisheries and aquaculture practices, such as genetic resources management and selective breeding.
In this review, we concentrated on 14 major fish species, considering their contribution to world fisheries and aquaculture production. These species are from diverse groups, including Cypriniformes (grass carp and common carp), Gadiformes (Atlantic cod), Perciformes (European sea bass, Nile tilapia, Asian sea bass, Pacific bluefin tuna and northern snakehead), Pleuronectiformes (tongue sole, turbot and Japanese flounder) and Salmoniformes (rainbow trout and Atlantic salmon). The grass carp, common carp and Nile tilapia are among the most important fish species in world aquaculture. European sea bass, Asian sea bass, Japanese flounder, rainbow trout and Atlantic salmon are vital species in aquaculture as well as fisheries. Atlantic cod and Pacific bluefin tuna are crucial species in marine fisheries.
This article – adapted and summarized from the original publication (Lu, G. and M. Luo. 2020. Genomes of major fishes in world fisheries and aquaculture: Status, application and perspective. Aquaculture and Fisheries, Volume 5, Issue 4, July 2020, Pages 163-173.) – summarizes potential genomic applications in fisheries and aquaculture that are related to assessment and use of genetic resources, disease resistance, growth and development, sexual determination, and fisheries management. It also discusses the challenges and perspectives of genomics in translational aquaculture and fisheries, which include genome assembly and annotation, genomic selection and breeding, genomics in fisheries management and integrated artificial intelligence systems.
https://www.aquaculturealliance.org/advocate/disruptive-technologies-for-aquaculture-part-1/
Genome sequencing and annotation
Genomics for aquaculture and fisheries made significant progress during the past decade since the first key fisheries species, Atlantic cod, was sequenced in 2011, and more fish species genomes are available. The genomic data in public repositories should, however, be used with caution because the quality of draft genomes varies greatly in different fish projects, which can be attributed to many factors such as the sequencing technologies and software tools used. Most currently published genomes were sequenced using second-generation sequencing technologies, which produce short reads that are challenging in assembly. The use of different sequencing technologies can fill this gap and produce chromosome-level, high-quality genomes.

The completeness of whole-genome sequencing refers to the percentage of the genome sequenced. In our review we noticed multiple assemblies submitted by different research groups existed in several fish species, so it is imperative to combine sequencing reads and come up with a reference genome for each species to establish a core set of universal single-copy genes in fish, which can be achieved using various bioinformatics tools. The resulting benchmark of single-copy genes will be instrumental for the assessment of genome assembly completeness and the study of fish systematics.
We propose to establish initiatives for functional annotation of fish genomes with a special interest in aquaculture or fisheries species. This community-based approach has been successfully demonstrated by several species, including zebrafish and salmonids, and such a community-engaged research initiative can avoid duplicated efforts in whole-genome annotation and functional gene validation.
There are dozens of important fisheries and aquaculture species with their draft genomes sequenced; however, the genomes of many other species are not available or require improvements. These species include silver carp (Hypophthalmichthys molitrix), bighead carp (H. nobilis), Catla (Catla catla), and Roholabeo (Labeorohita), which are ranked the 2nd, 5th, 7th and 10th in world aquaculture production, respectively. The draft genomes of invasive silver carp and bighead carp were reported; however, the improvement of genome assemblies and additional sequencing of native fish are needed. The genomic data for the important Indian carps Catla and Roholabeo remain limited in public databases.

Genomics and aquaculture
Molecular markers [molecules within a sample taken from an organism which can be used to reveal certain characteristics about the respective source] play an essential role in the selection and breeding programs in aquaculture and have been broadly used to construct the linkage maps [the linkage of genes in a chromosome] of important economic phenotypic traits such as growth, sex determination and pathogen resistance. The genetic gain [amount of increase in performance achieved through artificial genetic improvement programs] has been estimated to be greater than 12 percent per generation for growth rate and disease resistance through selective breeding in aquatic species.
Various researchers recently reviewed molecular marker-assisted breeding [used to help identify specific genes] in aquatic species and proposed a practical approach in selective breeding. In a selective breeding experiment with common carp, two sequential selections in a pool of more than 3 million individual mirror carp resulted in 300 phenotypically excellent individuals that were primarily from 15 families. The new stock grew more than 30 percent faster and exhibited superior genotypes [the complete set of genetic material of an organism] enriched by more than 140 percent compared to the control group. This genetic selection was conducted based upon only 20 molecular markers, which provides strong evidence of molecular marker-assisted selection (MAS) being a powerful approach in selective breeding.
Genomic selection estimates individual breeding values using a large number of markers distributed across the genome and has a high accuracy of selection that can lead to rapid increased genetic gain. Genomic selection occurs at the population level and thus reduces generation intervals by the selection of progeny based on genotypes. Importantly, genome selection can predict the breeding potential of candidate populations based upon phenotypic data, which is particularly suitable for the selection of economic traits that are difficult to measure or count. When considering the cost of genomic selection versus traditional progeny testing, the associated costs could be reduced by as much as 90 percent of the original cost. However, its use in aquaculture species (yellow croaker and Japanese flounder) has fallen behind compared to beef cattle and other livestock species.
Genome selection employs the prediction model based on genotypic and phenotypic data of a training population for the estimation of genomic estimated breeding values, GEBV [an animal’s unique DNA sequence to predict their true genetic merit more accurately] for all the individuals of the breeding population from their genomic profile. Fig. 1 shows a suggested pipeline for genomic selection and breeding in aquaculture, where the components related to different areas of science and technology were exemplified.
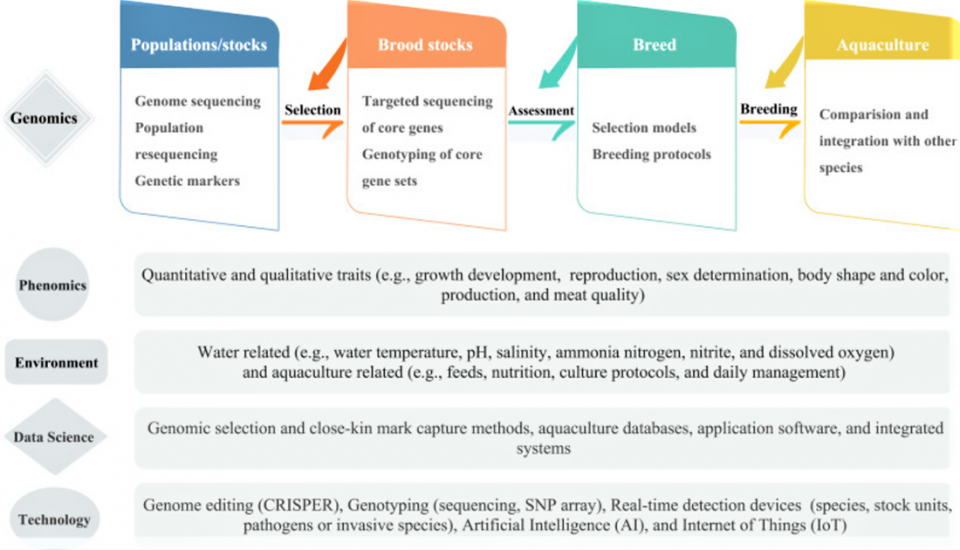
One of the challenges in the application of genome selection is the reliability of phenotypic data used for training prediction models. Several phenotyping techniques have been developed in plants using non-invasive imaging, spectroscopy, image analysis, robotics and high-computing facilities. These computer vision techniques have also been used in fish. Emerging technologies are needed for measuring the chemical (e.g., fat, protein, moisture) and physical (e.g., freshness, texture, color) attributes of fish with high accuracy. We anticipate future computer vision intelligent systems will be able to extract quantitative information from digital images more accurately and thus can increase the accuracy of phenotypic data, improving genome selection (Fig. 1).
Developing novel methods for the estimation of genomic breeding values (GEBV) has been quite active and developed to predict phenotypes from genotypes. For example, comparative analysis of different algorithms has been conducted to predict genetic values in large yellow croaker. Novel computing strategies were also developed for the prediction of GEBV, and the resulting computer program can be used by genomic selection programs. The inclusion of genotypic and environmental effects in genome selection models should result in a more precise selection in aquaculture practice (Fig. 1).

The whole-genome sequences offer a precious resource for molecular breeding through genome editing technologies. Genome editing technologies allow the interrogation of existing and novel genetic variation and thus facilitate the identification of causal genetic variation. For example, genome editing has been used to identify an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Genome editing and transgenic technology are essentially different. Unlike the introduction of foreign genes in transgenic organisms, genome editing can mutate particular positions of a targeted gene and therefore should be more easily accepted by consumers. The application of genome editing in aquaculture, although in its infancy, will undoubtedly become an essential means for the continued successful growth and stability of aquaculture production.
Genomics and fisheries
Genomics has brought new tools that can help address fundamental questions in fisheries management such as stock identification, population structure, and adaptive response to environmental change. The identification of single nucleotide polymorphisms [SNPs, pronounced “snips,” are the most common type of genetic variation – each SNP represents a difference in a single DNA building block, called a nucleotide] has enhanced the ability to trace fisheries products to their original locations, allowing regulation enforcement in some commercially important fish species.
Population genomic analysis through various advanced molecular techniques has been applied in several important fisheries species including Asian sea bass, and Atlantic cod which unveiled the genetic basis of fisheries-induced evolution and the potential effects of environmental change. However, such an effort needs to be expanded to more species that are important in world fisheries. Fisheries genomic research must focus on identifying informative SNPs that define management units and developing diagnostic markers for the monitoring of pathogens or invasive species.
Besides the identification of species and the origin of stocks, fish abundance and spawning stock biomass are important factors in fisheries management. The use of genetic markers to identify close-kin relationships provides fishery-independent estimates of spawning stock biomass. The close-kin approach has been applied to estimate the spawning stock biomass of Southern bluefin tuna. The close-kin method estimates the stock abundance based upon the genotypic data of sampled individuals, where the genotype of an individual can be considered a capture of the genotypes of each of its parents. However, this method requires a large number of samples in order to find sufficient numbers of parent-offspring pairs. Further evaluation of the close-kin method, including its use in the estimation of fish abundance, is needed in the future.

Applying genomic approaches to fisheries management is feasible and cost-efficient in most cases; however, the transformation of genomic findings into management practices has stagnated. The genomic tools and their power in species identification, determination of management units, and evaluation of natural resources shall give full consideration when fisheries management policies and guidelines are made. The stakeholders – including managers and fisheries geneticists – need to work collaboratively and make sure genomic tools become an integral component of fisheries management in the future.
Integrated and intelligent system for aquaculture and fisheries
Rapid advancements of next-generation sequencing technologies and broad interests in sequencing genomes of economically, ecologically, or evolutionarily significant species have made available hundreds of fish draft genomes in public repositories. Most fish genomes can be found in the genome database at the U.S. National Center for Biotechnology Information (NCBI), which has published the genomic data of 265 fish species, including 64 chromosome-level genome sequences (as of the end of 2019). Another important genome repository is Ensembl, which has made available approximately 60 fish genomes to the public.
However, the genome databases in NCBI and Ensembl are developed to serve diverse research communities and may not fulfill the needs of the aquaculture and fisheries community. In this respect, a few species-specific genome resources such as the Grass Carp Genome Database (GCGD) and the molecular data resource for salmonid species SalmoBase have been developed, which allows accessing genomic data, linkage mapping, and gene expression data. A database system dedicated to fish that integrates genetic, phenotypic, and environmental data is currently lacking.
It is thus important to develop an integrated big data platform for major species in aquaculture and fisheries. AgBioData is such a database system in agriculture, which could be adopted to aquaculture and fisheries to enhance genomics, genetics, and breeding research outcomes through standardization of protocols and practices. And moving forward with emerging technologies of data science and artificial intelligence (AI), the fisheries and aquaculture communities should strengthen collaborations and develop cloud sourcing projects to tackle challenging issues such as data sharing, integration and use in fisheries and aquaculture and promote technological innovations such as an Internet of Things, IoT, for Aquaculture 4.0.
Conclusions
The advances of next-generation sequencing technologies and genomics have revolutionized fisheries and aquaculture sciences and practices. We now have several dozens of important aquaculture and fisheries species with their complete genomes sequenced and available for analysis, comparison and knowledge discovery.
We have gained much knowledge about genomic mechanisms involved in germplasm resource utilization, disease resistance, growth and development, sexual determination and fisheries management. The full potential of genomic applications in genomic selection and fisheries management, however, has not been achieved.
In the coming decades, the application of genomic techniques such as genome editing and genomic selection, along with the use of emerging big data and artificial intelligence systems, are expected to leverage considerably sustainable breeding programs and achieve the goal of eradicating global poverty by 2030.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
Guoqing Lu, Ph.D.
Corresponding author
Department of Biology, University of Nebraska at Omaha
Omaha, NE 68182 USA -
Mingkun Luo, Ph.D.
Department of Biology, University of Nebraska at Omaha, Omaha, NE 68182 USA; and
Wuxi Fisheries College, Nanjing Agricultural University, Jiangsu, Wuxi, 214081, China



