The combined use of two genetic markers revealed that the majority of the products contained multiple species

Technological advancements in the seafood industry have supported the development of a broad range of appealing ready-to-eat and ready-to-cook products, which have significantly altered consumer perceptions of seafood. These innovative product forms are attracting new consumer targets that avoid seafood due to preparation time or a general dislike for fresh fish products, particularly among younger consumers. While some products, such as surimi or fish cakes, are clearly identified as mixtures of different species, others may disguise their composite nature under the appearance of higher-quality products like “fillets,” leading to misperceptions among consumers.
The diffusion of these new seafood forms not only boosts industry profits but also increases risks to consumers and marine ecosystems. But the assembly of mixed species in these products can facilitate illegal practices that compromise food safety and promote fraudulent activities involving species substitution. Indeed, such practices may pose health risks, particularly when allergenic species or specimens contaminated with harmful compounds, such as toxins or heavy metals, are introduced into the supply chain. Furthermore, illegal, unreported and unregulated (IUU) fishing activities contribute to the overexploitation of fish stocks and the degradation of marine ecosystems, compounding the ecological and economic challenges faced by global fisheries.
High-throughput sequencing technologies (HTS), based on Next Generation Sequencing (NGS) platforms, enable the simultaneous sequencing of all DNA molecules present and, therefore, the identification of every species in a sample. Despite its advantages, this approach has limitations that can significantly affect the results, particularly concerning the choice of molecular markers and primer pairs. These choices can influence PCR amplification, taxonomic profiles, and species-level resolution. To mitigate these biases, the use of multiple primer pairs from different molecular regions has been suggested for various applications, including the authentication of mixed seafood products and canned tuna.
This article – summarized from the original publication (Mottola, A. et al. 2024. Decoding Seafood: Multi-Marker Metabarcoding for Authenticating Processed Seafood. Foods 2024, 13(15), 2382 – presents the results of research that assessed the authenticity of processed single-species fish products using a multi-marker DNA metabarcoding approach.
Study setup
A total of 20 processed seafood samples (intact packages) from different brands and batches were collected from Italian supermarkets, including five burgers, four nuggets, three breaded fillets, three cutlets, three sticks and two products labeled as “fish fantasy.” The labels indicated that these products were made up of one (18 samples) or two species. The samples were transported to the food safety laboratory in an insulated bag. The cooled containers were maintained and stored at the proper temperatures until the time of molecular analysis.
Next Generation Sequencing, NGS (novel technology used for DNA and RNA sequencing) approaches are currently the most effective methods for verifying the accuracy of species declarations on processed seafood labels. Two primer pairs targeting the mitochondrial cytochrome c oxidase I, COI (the DNA barcoding system using the COI is very efficient for discriminating invertebrate and vertebrate species) and 16S rRNA genes (these gene sequences allow bacterial identification that is more accurate, robust and reproducible than from phenotypic testing) were sequenced. This analysis had three objectives: (i) evaluate the universality of the primer pairs utilized, (ii) assess the discriminatory power at the species level of the two sequenced fragments and (iii) compare molecular identifications with the list of ingredients reported on product labels.
Each product’s label was examined for compliance with Regulation (EU) No. 1169/2011 by checking the commercial name, ingredient list, net quantity, storage instructions, expiration date, manufacturer information, nutritional declaration and allergens. Molecular identifications were then compared with the species listed on the labels. Cases of mislabeling were identified if the species listed, either by scientific name or commercial designation, were not detected in the molecular analysis.
For detailed information on DNA extraction, amplification and sequencing; bioinformatics analysis; and label analysis and mislabeling assessment, refer to the original publication.
Can handheld DNA testing technology stand up to seafood fraud?
Results and discussion
In this study, authors used a multi-marker DNA metabarcoding approach to assess the species composition of processed seafood products that were presumably mono-species. Interestingly, the taxonomic profiles generated indicated that none of the products tested strictly adhered to a mono-species composition, and various species were either intentionally or accidentally included as ingredients. Furthermore, the analysis highlighted several limitations in the performance of the two primer pairs used, particularly in their ability to accurately trace products predominantly composed of Gadiformes (cods, hakes, pollock, haddock, and others) species.
The molecular analysis of 20 processed seafood samples using two DNA markers revealed the presence of 18 taxa spanning two classes, five orders, eight families and 11 genera. The taxonomic profiles generated by the COI and 16S markers were only partially overlapping, which underscores the need for a multi-marker approach in metabarcoding studies for food authentication, as supported by recent research.
Differences between the taxonomic profiles derived from the two primer pairs can be attributed to variations in the universality of the primers and their discrimination power at the species level. For instance, approximately 67 percent of sequences in the COI dataset were taxonomically assigned to the genus Gadus sp., followed by Merluccius sp. (28.3 percent), with other taxa detected in smaller proportions (overall < 1.5 percent). The 16S dataset showed about 60 percent of sequences assigned to Gadus sp., followed by Pleuronectidae (31.1 percent), with additional taxa detected in lesser quantities (overall < 6 percent).
This analysis not only confirmed four taxa shared between markers (Gadus sp., Salmo salar, Pollachius sp., and Pleuronectidae) but also identified seven taxa exclusive to the COI marker (Arctogadus glacialis, Merluccius sp., M. australis, M. hubbsi, M. paradoxus, Sander lucioperca, and Sebastes sp.) and five exclusive to the 16S marker (Chelidonichthys sp., Gadus chalcogrammus, G. morhua, Leptasterias coei and Limanda aspera) (Fig. 1).
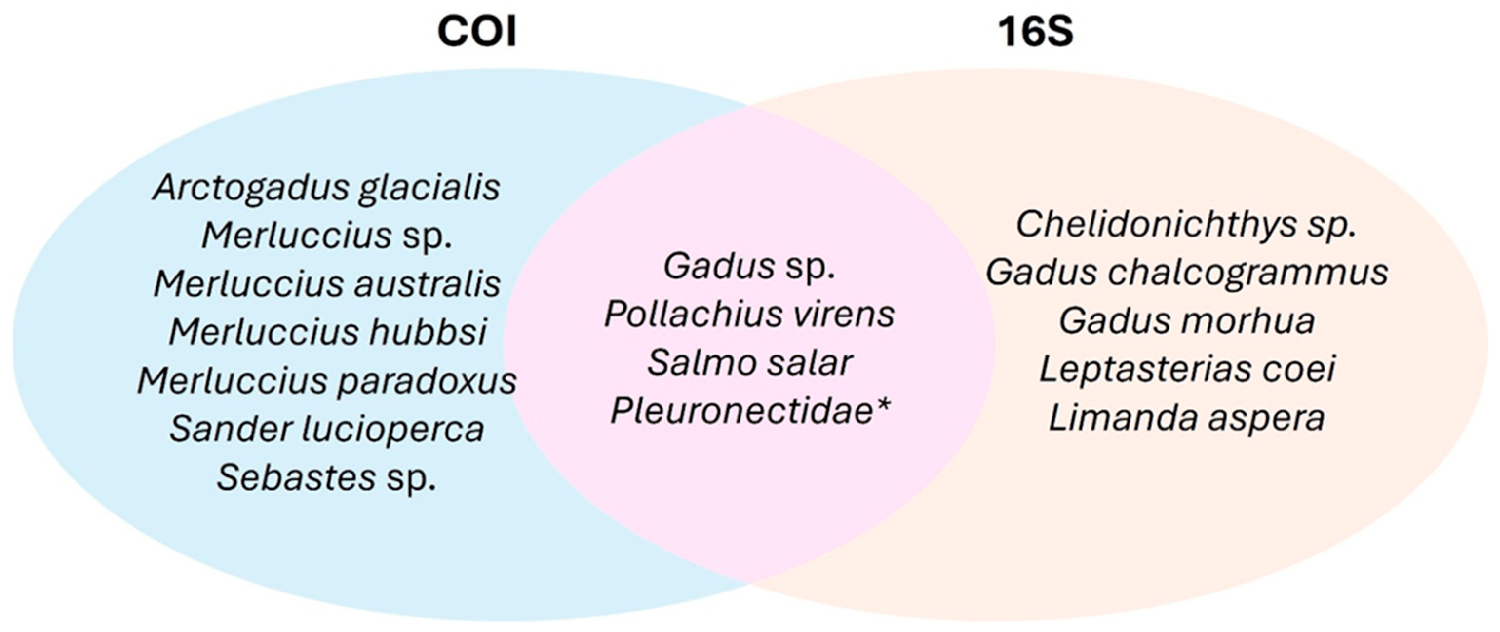
Although both primer sets detected the presence of the genus Gadus, they differed in their ability to discriminate between species within the genus. The COI marker did not differentiate between Gadus chalcogrammus and G. morhua, whereas the 16S marker effectively discriminated between them, assigning 36 percent of sequences to G. chalcogrammus and 16 percent to G. morhua. Additionally, the genus Merluccius was only detected by the COI marker, which successfully identified three species in this genus. However, four other species within this genus were not discriminated by this marker.
These results, pertaining to two commercially significant genera (Gadus and Merluccius) in terms of economic value and global fisheries and trade, underscore the critical importance of primer selection during experimental design in metabarcoding assessments. Furthermore, although both markers detected the presence of the Pleuronectidae family, only the COI marker could identify Pleuronectes platessa at the species level.
Regarding label analysis and product composition, the analysis of labels for all 20 processed seafood samples confirmed compliance with the mandatory requirements set by Regulation (EU) No. 1169/2011. Of these, 14 out of 20 labels (70 percent) voluntarily included both the commercial designation and the scientific name of the fish used. The remaining six labels (30 percent) provided only the commercial designation of the fish.
Based on the mislabeling criteria used, which defines mislabeling as the absence of molecular identification of the species declared on the label, the comparison of molecular identifications obtained by both markers with the information on the labels revealed that each product contained the declared species, at least at the genus level. This finding indicates a generally high level of compliance with voluntary declarations. However, two out of 20 samples (10 percent) were found to be mislabeled.
The observed substitution does not appear to be driven by economic motives, as hakes are generally considered higher-value species compared to pollock. Rather, the substitution may stem from confusion arising from the common names of these species, as both are referred to as “Merluzzi” in Italian. To mitigate such potential misunderstandings, which can occur among various intermediaries within the global supply chain, the adoption of Industry 4.0 technologies – such as artificial intelligence, blockchain, and big data – could enhance traceability “From Boat to Plate.” This integration promises to clarify product identities throughout the supply chain, thus supporting accurate labeling and compliance.
Although the products analyzed were labeled as containing one or two species, molecular analyses revealed that 95 percent of them were multi-species (Fig. 2), containing species not listed in the ingredients. These unexpected species included commercially recognized fish such as Pleuronectes platessa, Pollachius virens, Sander lucioperca, Merluccius australis, Sebastes sp., Limanda aspera, and Chelidonichthys sp., but also species not listed in the latest Italian regulations.
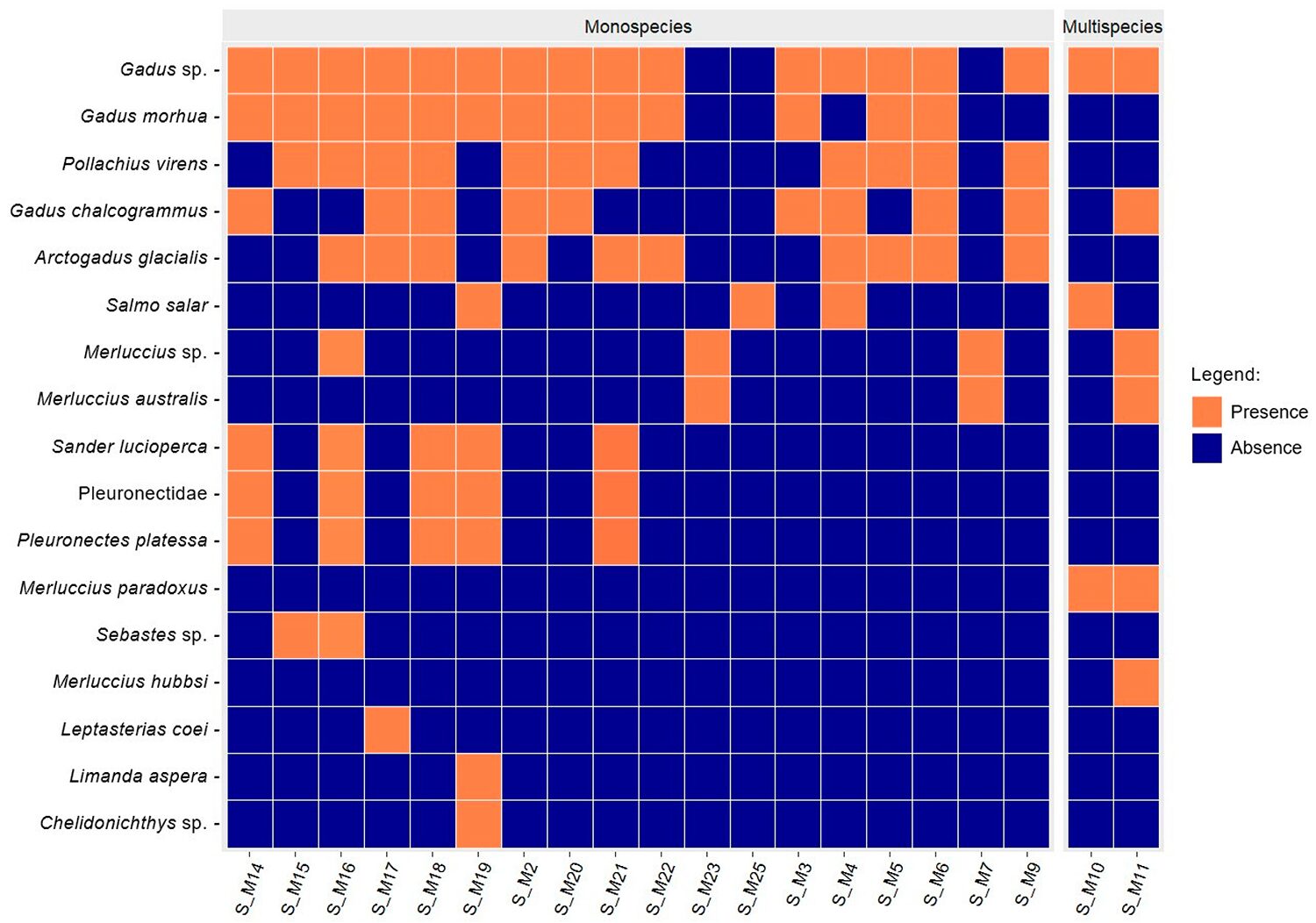
The presence of these species could be economically motivated, as the intentional addition of fish discards and low-value fish is a known industry strategy to increase product weight and boost profits. However, the presence of untraced seafood might also stem from illegal, unreported, and unregulated (IUU) fishing activities, raising serious ethical and ecological concerns. Moreover, such practices can pose risks to human health through unintentional exposure to heavy metals, persistent organic pollutants and microplastics.
In addition, the accidental inclusion of species could occur due to contamination along the seafood supply chain, particularly when different raw materials are processed within the same facility.
Perspectives
Overall, results confirm that the metabarcoding approach is a critical and effective tool for seafood traceability. Integrating molecular profiles obtained using multiple primer pairs has enhanced species identification success in seafood products. While both primer pairs used in this study exhibited limitations in tracing Gadiformes – the taxa most commonly used in these products – the COI fragments, considered the gold standard for metazoan (all animals having the body composed of cells differentiated into tissues and organs and usually a digestive cavity lined with specialized cells) assessments, performed marginally better.
Given the importance of primer selection, an optimal strategy for thorough species characterization in seafood could involve pairing one primer pair designed to amplify and distinguish species listed on product labels (expected species) with another primer pair that offers broader taxonomic coverage to detect unexpected species. Food fraud poses significant concerns for consumers, the food industry, and regulatory authorities, thus underscoring the need for innovative tools to ensure product authenticity.
Although additional species detected in our study were not categorized under mislabeling criteria, our findings suggest that truly mono-species products are rare in processed seafood. Voluntary declarations on labels often present a truncated list of ingredients, potentially used by brands as a marketing tactic to appeal to consumers seeking authentic, safe and wholesome products ostensibly made from a single species of fish. The urgency for suitable verification tools is clear. For highly processed matrices, NGS approaches stand as the sole effective method capable of tracing the full species composition of products.
Therefore, further development is necessary, particularly in laboratory setups and bioinformatic pipelines, before these methodologies can be routinely employed in official controls by authorities or within the food industry. However, their implementation will advance the “From Boat to Plate” strategy and combat fraudulent practices along the intricate fishery supply chain.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Author
-
Dr. Lucilia Lorusso
Corresponding author
Department of Veterinary Medicine, University of Bari Aldo Moro, Prov. le Casamassima 62, Km 3, 70010 Valenzano, Italy
Related Posts

Health & Welfare
Barcoding, nucleic acid sequencing are powerful resources for aquaculture
DNA barcoding and nucleic acid sequencing technologies are important tools to build and maintain an identification library of aquacultured and other aquatic species that is accessible online for the scientific, commercial and regulatory communities.

Intelligence
Japanese researchers deploy environmental DNA to understand biodiversity and solve aquaculture riddles
Can eDNA elicit insights into ocean biodiversity off the Ogasawara Islands, with implications for aquaculture monitoring?

Intelligence
DNA barcoding applications in aquaculture
DNA barcoding is an exciting tool for the identification of fish. Even non-taxonomists can classify organisms through the molecular taxonomy technique.

Fisheries
Can a data-sharing tool eliminate IUU fishing and make seafood supply chains more reliable?
The Sustainable Fisheries Partnership’s new data-sharing tool helps users identify environmental risks and eliminate IUU fishing in seafood supply chains.



