Detection by histopathology, and new in situ hybridization and PCR methods
Microsporidia are obligate intracellular parasites in a variety of hosts, from invertebrates to humans. Research for microsporidia has been more focused on terrestrial hosts, but approximately 50 percent of the known microsporidia genera infect aquatic hosts like crustaceans and fish. These microsporidian infections of aquatic hosts are considered a potential health hazard as well as a financial risk in aquaculture.
One of the critical diseases that impact shrimp aquaculture is “cotton shrimp disease” (CSD), and causative agents associated with this disease are microsporidia found in at least five genera including Pleistophora, Thelohania, Perezia, Agmasoma and Ameson. These microsporidian parasites mainly infect skeletal muscle, rendering the affected body regions white or opaque, which gives the common name to this disease. Lightly infected shrimp may look and behave normally, but heavy infections render the shrimp unmarketable or inedible. In shrimp, the white-opaque appearance of muscle is primarily linked to microsporidian infections. However, other causative agents such as dinoflagellates, bacteria, or viruses, may also be related to this disease.
This study focused on the microsporidium associated with CSD in shrimp collected from Madagascar, Mozambique and the Kingdom of Saudi Arabia. We investigated its histopathological characteristics and determined its taxonomical classification by small subunit rDNA (SSU rDNA) sequencing. These sequences are widely used for elucidating evolutionary relationships among organisms, since they are of ancient origin and are found in all known forms of life.
Also, specific diagnostic methods were developed for this disease based on: 1. in situ hybridization [ISH; a type of hybridization using a labeled complementary DNA, RNA or modified nucleic acids strand (i.e., probe) to localize a specific DNA or RNA sequence in a portion or section of tissue]; and 2. polymerase chain reaction (PCR), a method widely employed in molecular biology to make numerous copies of a specific DNA segment) targeting the SSU rDNA sequence.
Study setup
A total of 298 shrimp samples exhibiting clinical signs of CSD were submitted from Madagascar, Mozambique and Saudi Arabia to the Aquaculture Pathology Laboratory at the University of Arizona over a period of several years. Table 1 summarizes relevant information.
Han, CSD, Table 1
| Country and species | Life stage | Grade | Target organ | No. pos/total samples | PCR |
|---|
Country and species | Life stage | Grade | Target organ | No. pos/total samples | PCR |
|---|---|---|---|---|---|
| Madagascar (P. monodon) | Adult | G3-G4 | Muscle | 3/3 | NA |
| Madagascar (P. monodon) | Juvenile | G3-G4 | Muscle/HP | 6/30 | NA |
| Madagascar (P. monodon) | Sub-adult | NA | Muscle/HP | NA | pos. |
| Madagascar (P. monodon) | Sub-adult | G3-G4 | Muscle/HP | 4/19 | NA |
| Madagascar (P. monodon) | Adult | G2-G3 | Muscle/HP/heart | 3/5 | NA |
| Madagascar (P. monodon) | Sub-adult | G1-G3 | Muscle/HP | 6/28 | NA |
| Madagascar (P. monodon) | Juvenile | NA | Muscle | NA | pos. |
| Madagascar (P. monodon) | Juvenile | G3-G4 | Muscle/HP | 35/35 | NA |
| Madagascar (P. monodon) | Juvenile | NA | Muscle/HP | NA | pos. |
| Mozambique (P. monodon) | Juvenile | G3-G4 | Muscle/HP/gill/heart/LO | 4/23 | NA |
| Mozambique (P. monodon) | PL | G3-G4 | Muscle/HP/gill/heart/LO | 2/50 | NA |
| Mozambique (P. monodon) | Adult | G3-G4 | Muscle/HP/gill/heart/LO | 4/4 | NA |
| Saudi Arabia (P. indicus) | Adult | G4 | Muscle/HP/gill/heart/LO | 3/3 | NA |
| Saudi Arabia (P. indicus) | Adult | G2-G4 | Muscle/HP | 30/93 | NA |
| Saudi Arabia (P. indicus) | Adult | NA | Muscle | NA | pos. |
| Saudi Arabia (P. indicus) | Juvenile | G2-G4 | Muscle/HP | 5/5 | NA |
The Davidson’s alcohol-formalin-acetic acid (AFA) fixed samples were processed, embedded in paraffin and sectioned (4 µm thick) in accordance with standard methods (Lightner, 1996). After staining with hematoxylin and eosin (H&E) or Giemsa, the sections were analyzed by light microscopy and the severity of the infection was graded according to a pre-established G-grading system (from Lightner, 1996). Grading includes (G0) with no signs of infection/infestation by pathogen; (G1) signs of infection/infestation by pathogen are present, but at levels below those needed for clinical disease; (G2−G3) moderate to high signs of infection and/or infestation by number and severity of pathogen-caused lesions; and (G4) severe lesions and advanced tissue destruction with high pathogen numbers.
Results and discussion
Histopathology examination
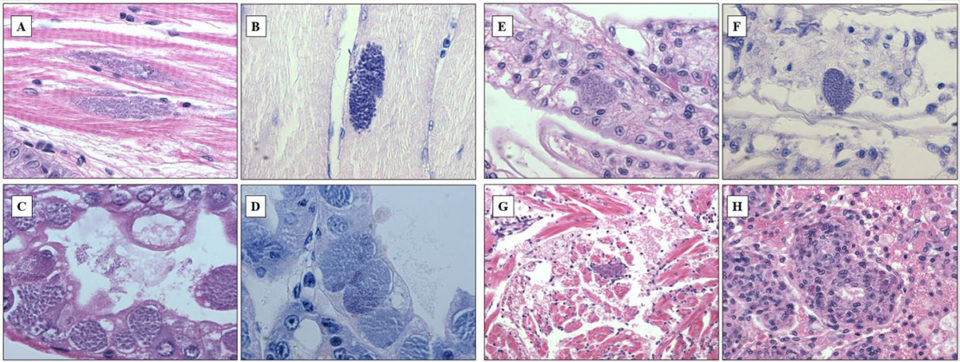
In shrimp and other crustaceans, Perezia spp. destroy the muscles and gradually replaces the musculature by masses of parasite developmental stages and spores. At the late stages of infection, parasites may invade other tissues and organs – such as cardiomyocytes, epithelial and connective tissue cells of the antennal glands – in the same shrimp. In this study, several tissues of the same shrimp were affected by this microsporidium. Histopathology examination revealed numerous microsporidian spores in samples collected from Madagascar, Mozambique and Saudi Arabia. The infection affected primarily hepatopancreas and muscle, at the severity level corresponding to grades ranging from G1 to G4.
Mature spores were seen among skeletal muscle fibers (Fig. 1A, B), and pre-spore stages and spores infected epithelium of hepatopancreas tubules (Fig. 1C, D). In severe infections, spores were also observed within epithelial cells of gill filament (Fig.1E, F), heart muscle fibers (Fig. 1G), and parenchymal cells of the lymphoid organ (Fig. 1H). Spores spreading all over the organism at the advanced stages of infection might have derived from muscle infection: muscles gradually deteriorated and spores contaminated surrounding tissues. In addition, most cases (90 percent) of CSD were accompanied by infections of gill fouling disease caused by Zoothamnium sp. or Epistylis sp., which indicated that animals suffering microsporidiosis were probably weak and susceptible to secondary infections or infestations.
SSU rDNA sequence of the new microsporidium causing CSD
We performed PCR targeting SSU rDNA sequences from the shrimp exhibiting CSD, collected in Madagascar, and a 1.2 kbp fragment was obtained and sequenced. The nucleotide sequence of SSU rDNA was deposited in Genbank. According to test results, the nucleotide sequence had a 94 percent identity to the SSU rDNA sequence of Pleistophora sp. infecting Penaeus setiferus, and 93 percent identity to that of an unidentified micro sporidium from Metapenaeus joineri. In 2002, Pleistophora sp. from P. setiferus was re-identified as Perezia nelsoni by morphological examinations. Based on sequence information, we therefore now consider the newly identified microsporidium to belong to the genus Perezia, and will refer to it as Perezia sp. hereafter. The CSD caused by Perezia sp. seems to be spread over several shrimp farming countries, including Madagascar, Mozambique, Saudi Arabia (the present study), the United States and Japan, but it has not been sufficiently studied.
In this study, extracted genomic DNAs from the Madagascar and Saudi Arabia samples were subjected to PCR with specific primers targeting SSU rDNA. The sequences obtained were 100 percent identical for both samples. Coincidentally, white spot syndrome virus (WSSV) detected in these same 3 countries (Madagascar, Mozambique and Saudi Arabia) also showed identical genomic patterns.
Perezia sp. – specific ISH
The probe strongly reacted to both skeletal muscle and hepatopancreas (Fig. 2A-C), and also other organs such as gill, heart, and lymphoid organ (data not shown), corresponding to the histopathology results. The probe designed from the SSU rDNA sequence of Perezia sp. appeared to be specific to shrimp with symptoms of CSD from Madagascar, Mozambique and Saudi Arabia.
At the beginning, we misidentified Perezia sp. infection as EHP infection by normal histopathology examination. However, this ISH probe did not react toP. stylirostris infected with E. hepatopenaei (Fig. 2D). No reaction was seen in any of the tissues prepared from pathogen-free P. vannamei. Also P. vannamei and P. monodon – as well as polychaetes infected with an unidentified Agmasoma-like microsporidium – did not react to ISH probes (data not included here).
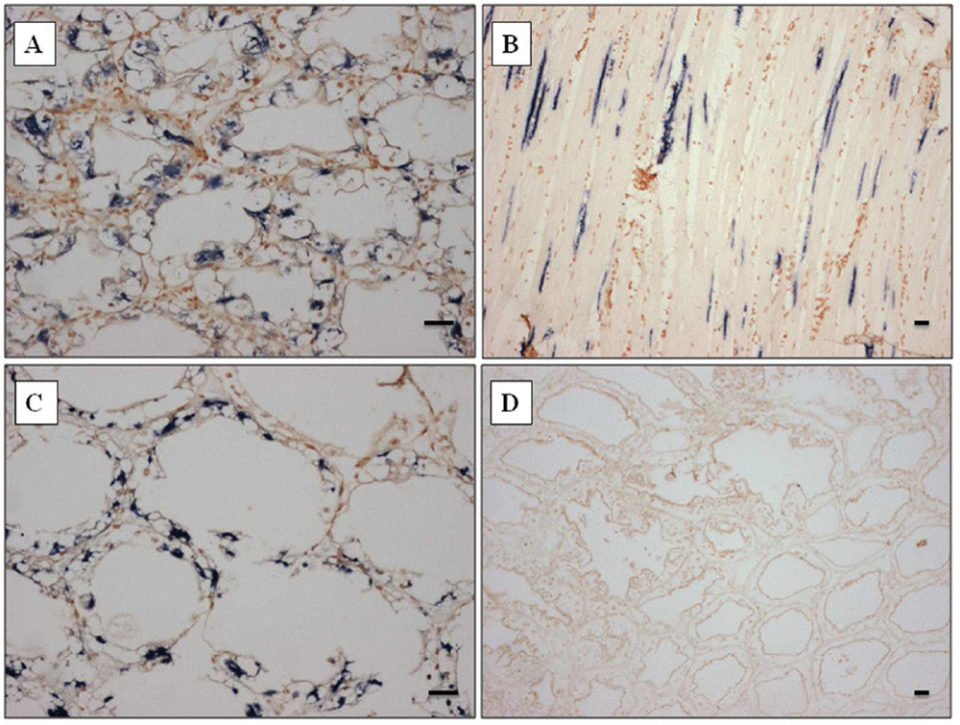
within hepatopancreas tubule epithelial cells of P. monodon from Madagascar. (D) Portion of the hepatopancreas from P. stylirostris from Brunei infected by Enterocytozoon hepatopenaei showing representative results, emphasizing the specificity of the probe as demonstrated by the absence of dark blue precipitate. Stain: Bismark-Brown counterstain. Scale bars = 30 μm
Perezia sp. – specific PCR
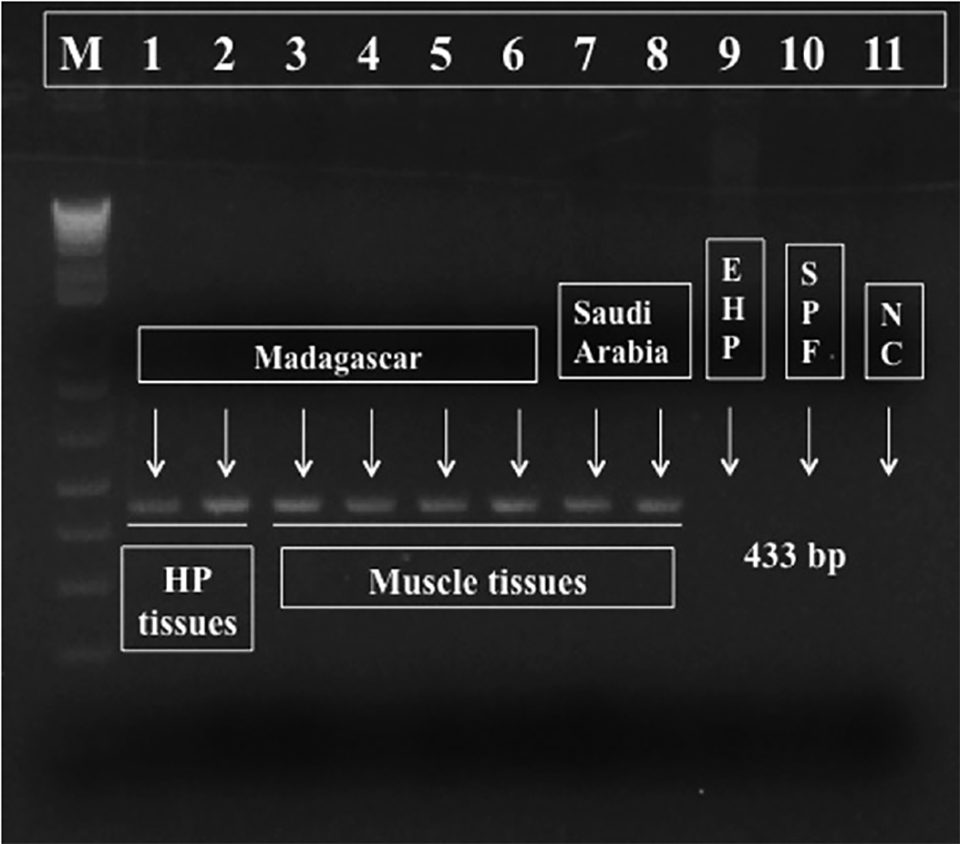
For diagnostic purposes, we designed a pair of PCR primers targeting Perezia sp. DNA and generated a 443 bp amplicon (molecular biology term to designate a piece of DNA or RNA that is the source and/or product of amplification or replication events), shown in Fig. 3. This PCR reaction is specific to the microsporidium Perezia sp. that causes CSD, and thus may be useful for monitoring this parasite. These primers did not react to the microsporidian DNA from E. hepatopenaei-infected shrimps (data not shown). Also, there was no cross-reaction to genomic DNA from shrimp (P. vannamei, P. monodon, P. indicus, P. stylirostris and Macrobrachium rosenbergii), polychaetes, squid and Artemia spp. (data not shown).
We did not analyze Mozambique samples by PCR, but the probe reacted intensely to ISH by depositing the reaction products in the target cells in Mozambique samples. In this study, we describe muscle and hepatopancreas-infecting microsporidia causing CSD in shrimps from Madagascar, Mozambique and Saudi Arabia in the Red Sea-Indian Ocean region.
Perspectives
Microsporidian infections were confirmed by histopathological analysis and the parasites were identified as Perezia sp. Also, we developed ISH and PCR assays capable of detecting this parasite. These methods can help shrimp producers in the diagnostic and management of CSD caused by the microsporidium Perezia sp.
References available from first author.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
Jee Eun Han, DVM, Ph.D.
Laboratory of Aquatic Biomedicine, College of Veterinary Medicine
Kyungpook National University, Daegu 41566, Korea -
Seung Chan Lee
Laboratory of Aquatic Biomedicine, College of Veterinary Medicine
Kyungpook National University, Daegu 41566, Korea -
Seul Chan Park
Laboratory of Aquatic Biomedicine, College of Veterinary Medicine
Kyungpook National University, Daegu 41566, Korea -
Marc Le Groumellec, DVM, Ph.D.
AQUALMA, Aquaculture de la Mahajamba
Mahajanga 401, Madagascar
Tagged With
Related Posts

Health & Welfare
Detection of an amoebic parasite in cultured Pacific white shrimp
This study describes an amoebic parasite infection in Pacific white shrimp cultured in an anonymous shrimp hatchery. The diseased shrimp exhibited grossly reduced appetite, lethargy, respiratory distress, eroded carapaces and blackened gills.

Health & Welfare
Assessment of supplemental Bacillus probiotics in whiteleg shrimp juveniles
Results of a feeding study supplementing probiotics in the diet showed that when the Bacillus species were complemented in an appropriate concentration into feeds, the growth and feed efficiency of whiteleg shrimp could be improved.

Health & Welfare
Potential applications of bacteriophages for AHPND control
Study demonstrates that isolated phages tested are effective in controlling AHPND infection in farmed penaeid shrimp and inhibiting bacterial growth.

Health & Welfare
Non-invasive diagnostic tool developed for shrimp disease EMS
The presence of AHPND-Vibrio parahaemolyticus can be detected both in fecal DNA samples and in the enriched bacterial broth with samples of enrichment broth showing increased sensitivity.


