Study shows how the gastrointestinal microbiome of shrimp varied between healthy and infected organisms

One of the most important diseases impacting shrimp-producing countries is Acute Hepatopancreatic Necrosis Disease (AHPND). The disease is caused by bacteria of the genus Vibrio-harboring pV-AHPND plasmids [small, extrachromosomal DNA molecules within a cell, physically separated from chromosomal DNA, and which can replicate independently] that encode genes for toxins. AHPND causes high mortality through a severe peeling of the hepatopancreatic cells, leading to the shrimp’s death. This mortality leads to considerable economic losses in the shrimp farming industry.
There is a trend towards reducing antibiotic use in aquaculture, and the use of probiotics – microorganisms providing beneficial effects to their hosts, either by preventing colonization of pathogenic bacteria through antagonism or promoting animal health through the stimulation of the immune system – is a significant, promising alternative to antibiotics and a way to treat and prevent cultured animal diseases. But there is still a need to identify beneficial groups of microorganisms among the normal resident microbiota and the mechanisms underlying the interactions with probiotics in the gastrointestinal tract, to efficiently use them to help improve the health of farmed shrimp.
The choice of probiotics can be controversial in some cases, particularly in aquaculture where certain probiotic genera are phylogenetically closely related to pathogens. For example, in Ecuadorian shrimp hatcheries, beneficial strains belonging to the bacteria species Vibrio alginolyticus and AHPND-causing strains of the species Vibrio parahaemolyticus belong to the same phylogenetic clade [a group of organisms composed of a common ancestor and all its lineal descendants], the Harveyi clade, and have been isolated from both healthy and non-healthy larvae cultures. In particular, a strain previously known as V. alginolyticus ILI strain [current genomic tools and available genomes allowed us to classify them as Vibrio diabolicus] has been isolated from healthy shrimp larvicultures and shown to be an efficient probiotic agent.
This article – adapted and summarized from the original publication [Restrepo et al. (2021). Microbial community characterization of shrimp survivors to AHPND challenge test treated with an effective shrimp probiotic (Vibrio diabolicus). Microbiome 9, 88 (2021) – reports on a study to evaluate the effect of the ILI strain and its potential to limit the detrimental effects of AHNPD using metabarcoding [the barcoding of DNA / RNA in a manner that allows for the simultaneous identification of many taxa within the same sample] on the gastrointestinal microbiota of shrimp after being challenged with AHPND-causing V. parahaemolyticus.
Study setup
We used the ILI probiotic strain provided by the Microbiology Laboratory at the Centro Nacional de Acuicultura e Investigaciones Marinas (CENAIM), previously identified as V. alginolyticus and characterized as an efficient probiotic. This strain was originally isolated from water of a healthy shrimp larvae hatchery and has been part of the CENAIM culture collection for more than 26 years.
After feeding the Pacific white shrimp (Litopenaeus vannamei) with the corresponding probiotic treatments, survival of the animals to a challenge with V. parahaemolyticusBA94C2 strain was evaluated.
A completely randomized design was applied, with four treatments and six replicates per treatment. Experimental units (40-liter tanks) were stocked with 40 shrimp (2.5 ± 0.3 grams). During the assay, shrimp were monitored for mortality every two hours. Differences of cumulative mortalities at 50 hours post-infection among infected treatments were analyzed, and shrimp survival recorded at the end of the experiment. Water exchange at 50 percent daily was done before and after the challenge, including during the period of probiotic feeding, but not the 50-hour challenge period. Shrimp were exposed to a 12-hour photoperiod per day.
For detailed information on the preparation of the probiotic inoculum, the immersion challenge test of shrimp supplemented with probiotics, and the several tests conducted, refer to the original publication.
Results and discussion
We evaluated the changes of the microbial community structure caused by AHPND on shrimp stomachs and hepatopancreas, with and without the presence of probiotics. Shrimp were either not challenged (healthy controls; C0) or challenged with AHPND-causing V. parahaemolyticus BA94C2 strain (AHPND-infected controls; C1) and compared with the same setup but with the daily supplementation of a probiotic (via feeding in a commercial feed without other probiotic medication) for a one-month period prior to the challenge. We used as probiotic the ILI strain (C2) and another probiotic (Bacillus sp. strain P64) for comparison (C3). The results from the challenge test showed the highest survival values for treatments C0 and C2 followed by C3. The experimental shrimp with the highest mortality were those of the AHPND-infected shrimp treatment (C1).
To date, there is very limited information on the effect of AHPND in the shrimp microbiome [characteristic microbial community occupying a reasonably well-defined habitat which has distinct physicochemical properties], and existing studies have focused on intestinal microbiota. Even though the contribution of intestinal microbiota on the impact of AHPND disease is not clear, we highlight the fact that the diversity on the intestine is higher than on the hepatopancreas but similar to the one found in sediment.
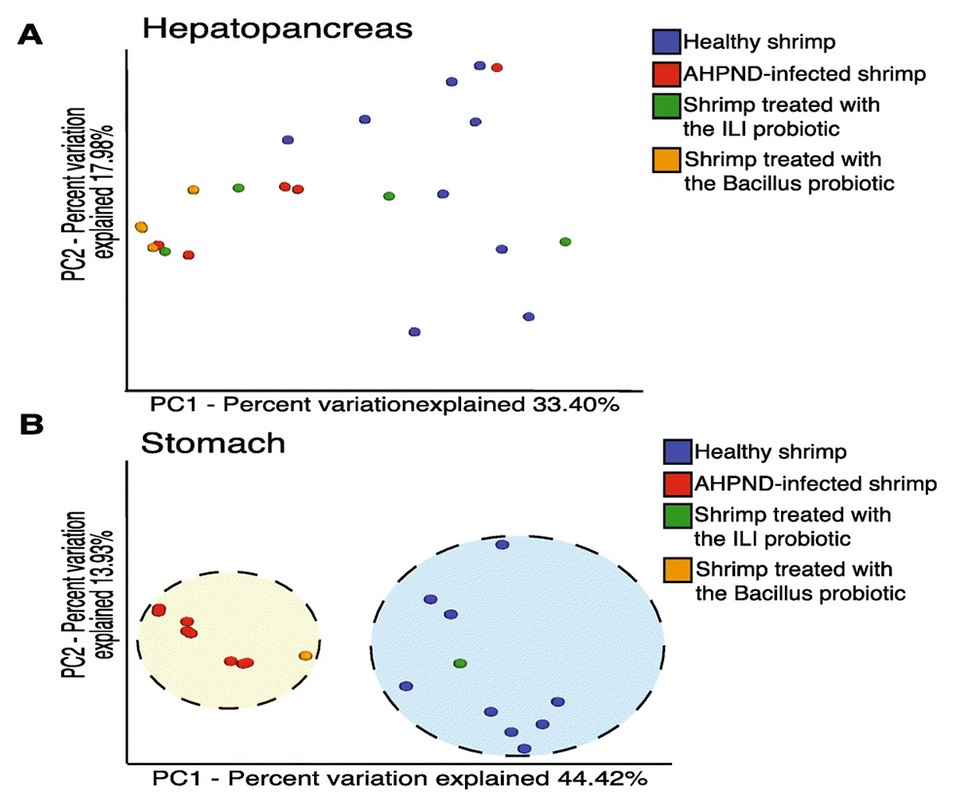
It is known that the immune-related organs – such as hemocytes, hepatopancreas and gills – are important in the response to AHPND disease. Also, the colonization of the stomach appeared to be the initial step of the infection for Vibrio-causing AHPND. Additionally, AHPND mainly affects the hepatopancreas and stomachs of infected shrimp; nevertheless, characterizing the microbiota from the shrimp’s stomach has not been the main focus to understand the changes in microbiota responses. Therefore, we examined microbiome alterations in response to pathogenic bacteria producing AHPND, in the stomach and hepatopancreas of shrimp.
We identified a significant loss of diversity in the microbiota of the hepatopancreas and stomach associated with AHPND. Our results contrast with some minimal effects observed in the intestinal microbiota of shrimp after White Spot Syndrome Virus (WSSV) infection and AHPND. Thus, we consider that the abundance and composition of the microbiota are influenced as a function of the organ in which it is located.
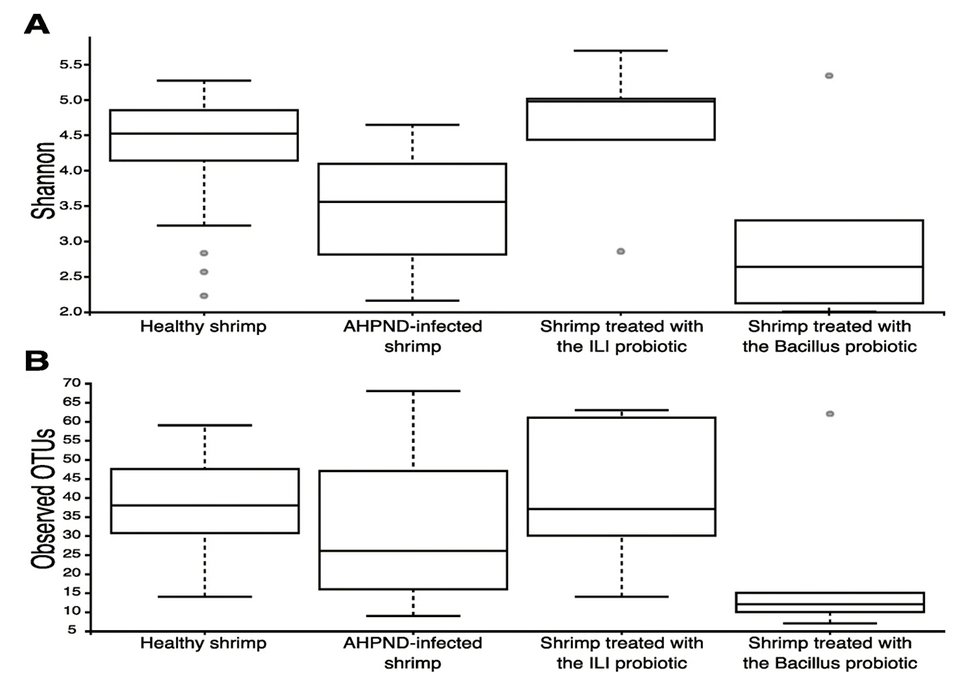
Interestingly, we identified members from the Tenericutes phylum, particularly those assigned to the genus Mycoplasmatales, as members of the healthy microbiome. Four amplicon sequence variants, ASVs [which refer to single DNA sequences recovered from a high-throughput marker gene analysis] from Mycoplasmatales belonged to the genus Mycoplasma, being the most abundant taxonomic group of the healthy stomach’s microbiome.
Various studies on the role of Mycoplasma spp. in other organisms have suggested that it would be harmless commensals [microorganisms living continuously on, or in certain parts of, the body, without causing disease] or endosymbionts [any organism that lives within the body or cells of another organism most often, though not always, in a mutualistic relationship] with the host; however, its specific role within the shrimp microbiome is unknown. Specific strains of Mycoplasma have been associated with healthy shrimp, potentially suggesting that it has a function in host health and recovery from disease states.
We were able to observe that the use of bacterial probiotics such as ILI can modulate the host microbiota and could contribute as a stimulant for several species that have probiotic potential by inhibiting gastrointestinal colonization of pathogens, as we found in this study.
The fact that ILI can acquire the causative plasmid for AHPND shows the high capacity of Vibrio species for Horizontal Gene Transfer, HGT [the movement of genetic material between unicellular and/or multicellular organisms other than by the (“vertical”) transmission of DNA from parent to offspring (reproduction)], which may be used to occupy different niches. The acquisition may vary in the aquatic environment and it may be influenced by the genetic background. The maintenance of these features in non-pathogenic bacteria could be evidence that secreted toxins are evolutionarily ancient features that may play a larger role in environmental fitness.
The ILI plasmid has not been described in any previous report, but it could provide important functions in the probiotic capacities that this strain exhibits. It is intriguing that no other reported pathogenic Vibrio strain contains a plasmid with similar gene content. The majority of large plasmids that are associated with Vibrio species have been characterized to be highly virulent against different hosts. Interestingly, we did not find any potential virulence factors encoded in the ILI plasmid; thus, the role of the plasmid in the bacteria is not yet clear. Further studies on the role of the plasmid in the ILI strain could clarify its function and potentially shed light on whether it contributes to the ILI’s probiotic capabilities.
Altogether, our data indicate that virulence is multifactorial in the Harveyi clade, and that the sources of virulence could be associated to specific hosts since many of the associated genes can be involved on the survival of the bacteria in different environments.
Perspectives
The use of probiotics in aquaculture is a practical alternative to promote animal health and prevent disease. We characterized the microbiome alterations generated by a successful probiotic that could control pathogenic populations in shrimp’s gastrointestinal tract and stimulate survival in aquaculture.
The V. diabolicus ILI strain is a bacterial strain isolated from the environment in a larval shrimp culture that has shown antimicrobial activity against many pathogenic strains of different Vibrio species. Supplementation of ILI to L. vannamei not only decreased the abundance of Proteobacteria amplicon sequence variants (ASVs) but also maintained a healthy-like microbial community in the shrimp gastrointestinal tract after being challenged with AHPND-causing bacteria.
Our findings suggest that the ILI strain can likely be used as a probiotic to reduce the population of pathogens for AHPND in the shrimp and to enhance survival and resistance against this important disease in shrimp aquaculture, without the risk of becoming a pathogenic organism.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
Leda Restrepo, M.Sc.
Department of Biological Sciences, Universidad de los Andes, Bogotá, Colombia; Max Planck Tandem Group in Computational Biology, Universidad de los Andes, Bogotá, Colombia; Escuela Superior Politécnica del Litoral, ESPOL, Centro Nacional de Acuicultura e Investigaciones Marinas, CENAIM, Campus Gustavo Galindo Km 30.5 Vía Perimetral, P.O. Box 09-01-5863, Guayaquil, Ecuador
-
Cristóbal Domínguez-Borbor, M.Sc.
Escuela Superior Politécnica del Litoral, ESPOL, Centro Nacional de Acuicultura e Investigaciones Marinas, CENAIM, Campus Gustavo Galindo Km 30.5 Vía Perimetral, P.O. Box 09-01-5863, Guayaquil, Ecuador
-
Leandro Bajaña
Escuela Superior Politécnica del Litoral, ESPOL, Centro Nacional de Acuicultura e Investigaciones Marinas, CENAIM, Campus Gustavo Galindo Km 30.5 Vía Perimetral, P.O. Box 09-01-5863, Guayaquil, Ecuador
-
Irma Betancourt
Escuela Superior Politécnica del Litoral, ESPOL, Centro Nacional de Acuicultura e Investigaciones Marinas, CENAIM, Campus Gustavo Galindo Km 30.5 Vía Perimetral, P.O. Box 09-01-5863, Guayaquil, Ecuador
-
Jenny Rodríguez, Ph.D.
Escuela Superior Politécnica del Litoral, ESPOL, Centro Nacional de Acuicultura e Investigaciones Marinas, CENAIM, Campus Gustavo Galindo Km 30.5 Vía Perimetral, P.O. Box 09-01-5863, Guayaquil, Ecuador; Escuela Superior Politécnica del Litoral, ESPOL, Facultad de Ciencias de la Vida, FCV, Campus Gustavo Galindo Km 30.5 Vía Perimetral, P.O. Box 09-01-5863, Guayaquil, Ecuador
-
Bonny Bayot, Ph.D.
Escuela Superior Politécnica del Litoral, ESPOL, Centro Nacional de Acuicultura e Investigaciones Marinas, CENAIM, Campus Gustavo Galindo Km 30.5 Vía Perimetral, P.O. Box 09-01-5863, Guayaquil, Ecuador; Escuela Superior Politécnica del Litoral, ESPOL, Facultad de Ingeniería Marítima y Ciencias del Mar, FIMCM, Campus Gustavo Galindo Km 30.5 Vía Perimetral, P.O. Box 09-01-5863, Guayaquil, Ecuador
-
Alejandro Reyes, Ph.D.
Corresponding author
Department of Biological Sciences, Universidad de los Andes, Bogotá, Colombia; Max Planck Tandem Group in Computational Biology, Universidad de los Andes, Bogotá, Colombia; Center for Genome Sciences and Systems Biology, Department of Pathology and Immunology, Washington University in Saint Louis, Saint Louis, MO, USA


