Study concludes that the currently farmed P. vannamei lines in Ecuador are tolerant to circulating IHHNV genotypes

The Infectious Hypodermal and Hematopoietic Necrosis Virus (IHHNV) has been listed as a notifiable crustacean pathogen by the World Organization for Animal Health (OIE) since 1995 and is highly prevalent in America, Asia and Australia. The virus previously caused large-scale mortalities in Pacific blue shrimp (Penaeus stylirostris) and causes growth retardation called runt-deformity syndrome (RDS), in Pacific white shrimp (Penaeus vannamei) and black tiger shrimp (Penaeus monodon). Currently, the presence of five genotypes (three infectious types: I, II, and III, and two non-infectious types: A and B) of IHHNV has been documented.
IHHNV is an endemic pathogen in most Latin American countries including Ecuador, Mexico, Honduras, Colombia, Brazil and Peru. Infection with IHHNV has been reported in several life stages of P. vannamei, including postlarvae, juveniles and broodstock, without any clinical signs, including RDS. At the farm level, the prevalence of the virus can range between 10 to 50 percent, and the animals may appear healthy. However, it remains unknown whether the presence of the virus makes shrimp more susceptible to other pathogens and whether chronically infected animals have an impact on growth and farm productivity.
Updates on the current impact of IHHNV on the Ecuadorian and Peruvian shrimp industry are insufficient to determine the pathogen impact at the farm level. According to the OIE, however, from 2012 to 2019 there have been total of 25 and 21 cases reported of IHHNV in grow-out ponds in Ecuador and Peru, respectively.
This article – adapted and summarized from the original publication [Aranguren Caro, L.F. et al. 2022. Current status of infection with infectious hypodermal and hematopoietic necrosis virus (IHHNV) in the Peruvian and Ecuadorian shrimp industry. PLoS ONE 17(8): e0272456] – reports on a study to determine the influence of the circulating infectious strains(s) of IHHNV present in Peru and Ecuador on shrimp production in these two countries using three different approaches. These include 1) determining the prevalence of IHHNV and assessing the effect of IHHNV on shrimp growth in commercial grow-out ponds; 2) determining the full-length genome sequence of IHHNV genotypes in Ecuador and Peru and examining their relationship to other IHHNV isolates reported elsewhere in the world; and 3) assessing the infectivity of Peru and Ecuador isolates of IHHNV on three different penaeid species, P. vannamei, P. monodon and P. stylirostris.
Study setup
The IHHNV isolates were collected from Tumbes and Piura, north of Peru, and three regions in Ecuador, namely El Oro, Guayaquil and Esmeraldas. Sampling was conducted by the National Fisheries Health Agency in Peru (SANIPES) and the National Aquaculture Chamber in Ecuador.
Six IHHNV isolates from Peru were collected between 2019 and 2020 from shrimp farming regions in Tumbes and Piura. In Ecuador, grow-out ponds displaying a high coefficient of variation (CV percent >20) with a history of IHHNV occurrence were selected in 2020. Eight grow-out ponds distributed in Guayas, El Oro and Esmeraldas were analyzed. From each grow-out pond, 30 shrimp were randomly sampled, weighed and had their pleopod and gill tissues preserved for real-time PCR analysis. Additional shrimp collected from each of these locations were individually frozen (minus-20 degrees-C) for further analysis. Finally, out of the same pond, 5 to 10 shrimp were fixed in Davidson’s fixative for histological analysis. Samples from Peru and Ecuador were sent to the University of Arizona Aquaculture Pathology Laboratory (UA-APL) for further analysis.
For detailed information on IHHNV detection and genome sequencing; phylogenetic analyses; the IHHNV challenge test; histopathology and in situ hybridization; and statistical analyses, refer to the original publication.
Testing may help increase IHHNV tolerance in Pacific white shrimp
Results and discussion
Here we present data on the prevalence of IHHNV and its impact on the growth of P. vannamei in grow-out ponds in three major shrimp-producing regions in Ecuador. We also determined the genome sequence of the virus in P. vannamei originating in Ecuador and its neighboring country, Peru, where IHHNV is prevalent. Finally, we performed an experimental bioassay using IHHNV isolates originating from Ecuador and Peru to determine the virulence of the virus on three SPF penaeid shrimp species: P. vannamei, P. monodon, and P. stylirostris.
Farm analysis
Samples of P. vannamei from grow-out ponds representing three different regions in Ecuador were screened for IHHNV. The virus was present in samples collected from all eight grow-out ponds located in the three different regions, and the IHHNV prevalence varied widely from 3.3 up to 100 percent. In real-time PCR analysis, some shrimp showed negative results, whereas others showed a copy number of up to 2.9 × 105 copies/ng DNA.
To establish the association between shrimp weight and the presence of IHHNV, the population from each pond was divided into two groups based on real-time PCR results (IHHNV-positive and IHHNV not detected). As shown in Fig. 1, there were no significant correlations between the average shrimp weight and the presence of IHHNV within the same pond.
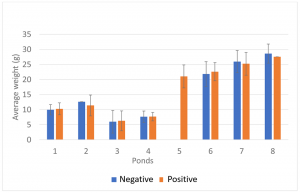
Sellars, et al., showed that in a P. monodon farm in Australia, growth performance and survival were reduced in grow-out ponds with a high copy number of IHHNV (0.5 × 103 to 2.24 × 106 copies/ng DNA). In our study, the mean copy number of IHHNV varied between 5.9 × 100 and 1.4 × 104 copies/ng DNA, which is lower than the IHHNV load reported by Sellars, et al., which may partially explain the little to no influence on growth performance. There was a significant direct correlation between the IHHNV copy number and the prevalence of IHHNV in the grow-out ponds. The prevalence and copy number was high. Similar results in grow-out ponds were reported by Sellars et al. These findings are consistent with the pattern of infection with infectious agents.
To better understand the effect of IHHNV load on the growth of P. vannamei shrimp in a given pond, shrimp populations were divided into three categories based on the IHHNV copy number: not detected, low to medium, and high. Only three out of eight grow-out ponds (ponds 2, 3 and 7) had shrimp representative of all three categories. As shown in Fig. 2, there was an inverse trend between the IHHNV copy number and shrimp weight, with animals having higher viral loads and lower weights. In one case (pond 2), a significant correlation was observed. These grow-out ponds had the highest mean copy numbers of IHHNV. One grow-out pond (pond 5) that also showed a high IHHNV mean copy number was not analyzed owing to the fact that 100 percent of the shrimp tested positive for IHHNV. It is worth noting that RDS was not observed in shrimp collected from any of the three ponds or the remaining five ponds.
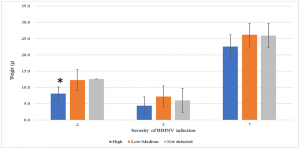
* Represents a significant difference by one-way ANOVA (P<0.05). Adapted from the original.
Historically, IHHNV was reported to cause RDS in P. vannamei, resulting in large size disparities and growth deformities. A recent report from India documented deformities, including deformed sixth abdominal segment, deformed rostrum, cuticular roughness and wrinkled antennae, and a wide variation in the size of grow-out ponds in P. vannamei analyzed from 350 shrimp farms. In that study, unfortunately, only shrimp with clinical signs were analyzed; therefore, it was not possible to determine IHHNV prevalence in healthy individuals from the same ponds. An earlier study from India reported the presence of two pathogens, White Spot Syndrome Virus (WSSV) and IHHNV, without any external deformities. From this perspective, our findings suggest a unique first glimpse of the inherent ability of certain genetic lines of P. vannamei to carry a high viral load without displaying any clinical signs.
IHHNV genome sequence and phylogenetic analyses
In our study, the full-length IHHNV genome sequence (AF218266) was used as a reference for mapping. The IHHNV genome varies in length; for example, the IHHNV isolated from Brazil is 3739 nt in length [a nucleotide, abbreviated nt, is a common unit of length for single-stranded nucleic acids], and the IHHNV isolated from India is 3908 nt in length. In our study, the IHHNV genome had an average size of 3957 nt, which was in the range of the IHHNV genomic size. Also, through various analyses, we demonstrated the uniqueness of the novel IHHNV sequences found in Latin America.
The phylogenetic tree [diagram showing the evolutionary relationships among various biological species] we constructed from the full genome sequence of IHHNV revealed that all IHHNV isolates from Peru and Ecuador belonged to the same clade type 2 IHHNV group as IHHNV isolates from East Asia and the Americas, as described elsewhere. However, it seems that they form a new sub-clade [a clade is a group of organisms composed of a common ancestor and all its lineal descendants] within this type 2 strain. Therefore, we propose a new type of IHHNV isolated from these two geographical regions. One possible explanation is the frequent movement of postlarvae from Ecuador to Peru. During active surveillance conducted by SANIPES, samples of imported postlarvae from Ecuadorian lots were analyzed by PCR. The frequency of IHHNV in these samples was 55 percent (15 out of 27 samples) in 2019, 52 percent (75 out of 144 samples) in 2020, and 76.3 percent (55 out of 72 samples) in 2021.
Virulence of IHHNV isolates from Ecuador and Peru determined by experimental challenge
For the experimental challenge, IHHNV isolates from Ecuador and Peru were used to experimentally infect SPF shrimp. The virulence of the IHHNV isolates was determined based on the final survival rate, RDS, IHHNV copy number, histological lesions and in situ hybridization (ISH). Although SPF P. vannamei were used for the IHHNV bioassays for both the Ecuador and Peru isolates, two additional species, P. monodon and P. stylirostris, were also tested for the IHHNV-Ecuador bioassay. Overall, the survival was very high in all three species (97–100 percent survival), even when the inoculum was administered via injection with a high viral copy number (9.0 × 106 copies). These results clearly support the notion that these SPF lines are tolerant to the IHHNV isolates used in the bioassay.
Despite the lack of mortality in the bioassay, IHHNV-injected shrimp had a high IHHNV load at the end of the experiment. The IHHNV strains from Peru and Ecuador did not cause mortality in two endemic shrimp species (P. vannamei and P. stylirostris) and one exotic shrimp species (P. monodon). These findings can be explained in part by considering the historical background of the P. stylirostris stock. The genetic line of P. stylirostris used in this study originated from Mexico and is likely a progenitor of an IHHNV-resistant stock that was selected from survivors of an IHHNV outbreak.
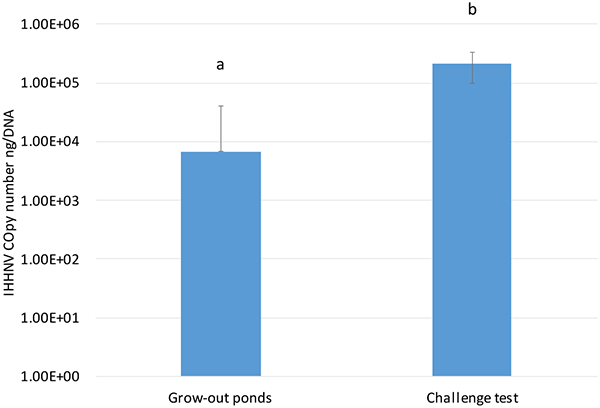
When the IHHNV load in P. vannamei raised in grow-out ponds in Ecuador was compared to the IHHNV load found in the SPF P. vannamei used in the experimental challenge, there was a significant difference in the viral loads (P<0.001), with a higher IHHNV copy number in the SPF population used for the challenge test than in the farm-raised P. vannamei (Fig 3). Although the genetic backgrounds of the SPF line used in the bioassay and the farm-raised P. vannamei are unknown, the data suggest a high tolerance of endemic shrimp populations that are being raised in the Ecuadorian shrimp industry. Currently, in Latin America, most shrimp have been raised under conditions where IHHNV is endemic, and the use of pond-rear broodstock has apparently acquired unintentional selection for resistance to diseases, including IHHNV. This tolerance/resistance may explain the absence of RDS associated with IHHNV infection in commercial grow-out ponds rearing P. vannamei.
In Latin America’s shrimp farming industry, IHHNV has been one of the most prevalent pathogens reported in the last 20 years by the World Animal Health Information System (WAHIS). Viral accommodation is a hypothesis proposed earlier in 2009 and then updated in 2019. It is related to tolerated persistent infection where viruses or fragments of their genome are inserted into the shrimp genome by an independent host mechanism that might have a protective effect, which could explain the absence of clinical signs. Furthermore, Taengchaiyaphum, et al., recently confirmed a significant reduction in the IHHNV copy number in P. vannamei where IHHNV circular viral copy (cvcDNA) was present. This phenomenon could explain the absence of RDS and severe histological lesions in shrimp infected with IHHNV in Latin America.
New circular DNA vaccines and heritable, anti-viral immunity possible for shrimp
Conclusions
IHHNV is a pathogen with high prevalence in shrimp farms in Latin America. We surveyed the prevalence of IHHNV in commercial grow-out ponds in three major shrimp-producing regions of Ecuador and assessed its impact on shrimp growth. Upon confirming the presence of the virus in commercial ponds in Ecuador, we determined the genome sequence of the virus in its endemic range from Ecuador and Peru. Finally, using an experimental bioassay, we determined the virulence of the IHHNV-Ecuador and IHHNV-Peru isolates using three different penaeid shrimp species: P. vannamei, P. monodon and P. stylirostris. Our data revealed that IHHNV did not cause mortality in the challenged species and did not have a noticeable impact on shrimp production.
A wide variation in IHHNV prevalence is likely due to many factors, such as the days of culture when the samples were collected, health status of the postlarvae used to stock the ponds, genetic background of the postlarvae, culture conditions, farm management and other unknown factors. Leaving aside the percent prevalence data, in ponds with animals carrying no IHHNV to high levels of IHHV, we found a negative trend between the IHHNV load and shrimp weight in the corresponding pond. Next-generation sequencing (NGS) data revealed that circulating IHHNV genotypes in Ecuador and Peru represent the infectious form of IHHNV.
Finally, in laboratory bioassays that lasted 30 days using IHHNV isolates from Ecuador and Peru, we could not reproduce any clinical manifestation of IHHNV infection in P. vannamei or any significant mortality in any of the three penaeid shrimp species. In addition, although IHHNV was detected in all three species using real-time PCR, the viral load was significantly higher in P. vannamei than in P. monodon and P. stylirostris. Moreover, using H&E histology analysis, Cowdry type A inclusion bodies – which are considered distinctively characteristic of a particular disease – of IHHNV infection, were detected in P. vannamei and P. monodon, but not in P. stylirostris. In situ hybridization (ISH) was performed to delineate the IHHNV-specific lesions in P. vannamei and P. monodon.
Although our data on IHHNV quantitative load and its effect on growth are limited, they suggest a negative correlation between these two parameters. Therefore, a more robust data set with a larger number of samples coupled with the health status of the postlarvae used to stack a pond will be immensely beneficial for a more accurate investigation of a quantitative relationship between shrimp growth and pathogen load of circulating IHHNV genotypes in Latin America.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-

Luis Fernando Aranguren Caro, Ph.D.
Corresponding author
Aquaculture Pathology Laboratory, School of Animal and Comparative Biomedical Sciences, The University of Arizona, Tucson, Arizona USA -
Muriel Maria Gomez-Sánchez, MVZ
Subdirección de Sanidad, Dirección de Sanidad e Inocuidad (National Fisheries Health Agency in Peru, SANIPES), San Isidro, Lima, Perú
-

Yahira Piedrahita, M.Sc.
Cámara Nacional de Acuacultura, CNA, Guayaquil, Ecuador
-

Hung Nam Mai, Ph.D.
Aquaculture Pathology Laboratory, School of Animal and Comparative Biomedical Sciences, The University of Arizona, Tucson, Arizona USA
-
Roberto Cruz-Flores, Ph.D.
Aquaculture Pathology Laboratory, School of Animal and Comparative Biomedical Sciences, The University of Arizona, Tucson, Arizona, USA; and Centro de Investigación Científica y Educación Superior de Ensenada (CICESE), Ensenada, Baja California, México
-

Rod Russel R. Alenton, Ph.D.
Aquaculture Pathology Laboratory, School of Animal and Comparative Biomedical Sciences, The University of Arizona, Tucson, Arizona, USA
-

Arun K. Dhar, Ph.D.
Aquaculture Pathology Laboratory, School of Animal and Comparative Biomedical Sciences, The University of Arizona, Tucson, Arizona, USA
Tagged With
Related Posts

Health & Welfare
A study of Zoea-2 Syndrome in hatcheries in India, part 1
Indian shrimp hatcheries have experienced larval mortality in the zoea-2 stage, with molt deterioration and resulting in heavy mortality. Authors investigated the problem holistically.

Health & Welfare
Challenging Pacific white postlarvae with AHPND
Study results indicate that P. vannamei challenged with AHPND in biofloc had higher survival rates than shrimp challenged in clear water.
Health & Welfare
Emerging disease: Shrimp Hemocyte Iridescent Virus (SHIV)
SHIV is a new Pacific white shrimp virus in the Iridoviridae family. Authors also developed an ISH assay and a nested PCR method for its specific detection.

Health & Welfare
Measuring susceptibility differences to IHHNV infection
This study evaluated the susceptibility to IHHNV in three different shrimp batches by measuring infectivity titers. Each batch showed a different infectivity titer, therefore, each batch had a specific susceptibility to IHHNV.



