Micro-satellites are essential tools to develop genetic maps and identify quantitative trait loci

The Atlantic halibut (Hippoglossus hippoglossus) has promise to become a commercially important marine fish species for aquaculture. As with most marine fish currently in the process of domestication, Atlantic halibut production still primarily relies on the spawning of fish caught in the wild. Only in the last few years have captive mature broodstock been available. Ideally, selection programs will identify the best performers of these first-generation domestic broodstock to develop a strain of fish that is better suited to aquaculture conditions and has improved performance.
Random broodstock selection
In 1996, the Canadian halibut aquaculture industry was still in its infancy, but the relatively long time (5 to 7 years) to sexual maturity of these fish was a key factor in the realization that the development of a program for selecting domesticated broodstock was needed sooner rather than later. Other limitations faced by the nascent broodstock selection program included the limited number of broodstock that could be carried due to animal size, feed costs, and the need to combine families of offspring into the few available tanks.
Consequently, the initial selection of broodstock was effectively random. It came from a pool of fish produced from 80 crosses between 14 females and 13 males that were carried out that spawning season. One hundred forty-five F1 fish were chosen as potential future broodstock.
Genetic relationships
As these fish approached sexual maturity, it became apparent that the relationships among the animals needed to be sorted out. The authors chose to use genetic markers called micro-satellites to determine the pedigrees of the fish. Microsatellites are regions of DNA that contain a short (2 bp in this case) sequence that is repeated 20 or more times in a row.
Microsatellites are highly variable among individuals in a population, yet are inherited by offspring from their parents. Using a set of five microsatellite markers, the work exactly identified the parents of 142 fish, while for the other three, one parent could not be resolved between two possibilities.
Limited genetic pool
The data was then compared to the spawning records for 1996 and it became apparent that several of the 1996 crosses were highly represented among the retained fish (Fig. 1). In fact, more than half the fish came from only four crosses: numbers 1, 41, 49, and 51. The majority (64 percent) of the 1996 crosses, however, were not represented in the retained fish. The offspring of three of the original male parents and four of the original female parents were completely absent from the retained group.
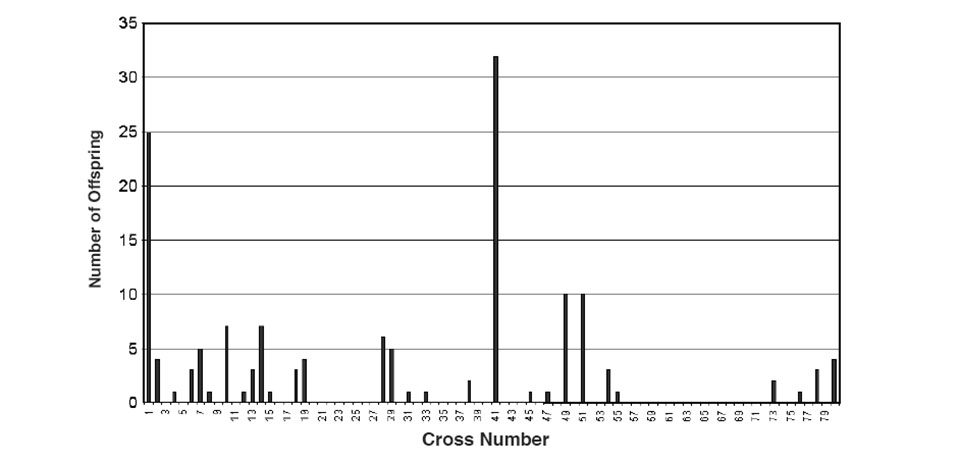
On a genetic level, the total number of alleles in the F1 fish was about 75 percent of the original parents. Statistical analysis indicated the effective population size or number of genetically different contributors of the F1 fish was less than half the initial size of 27 (14 females and 13 males). Clearly the random selection of the offspring of these fish had significant consequences on the depth of their genetic pool.
Need for genetic monitoring
These results demonstrated the need to begin genetic monitoring as soon as any selection, intentional or not, is made. In this case, the initial selection had a substantial effect on the potential genetic variation available to the domestication program.
The high degree of variability in the success of these crosses can be attributed to the use of hatchery techniques that were under development, but more even representation between crosses can be expected with improvements in culture technique. However, the potential for future inbreeding among the F1 fish, especially in the absence of any genetic monitoring, has become much greater.
With more than half the F1 fish coming from four families, it is very likely that crosses of randomly chosen fish would result in mating between siblings, which, if repeated in future generations, would lead to a highly inbred line with little genetic variation and therefore little opportunity to improve performance. However, future crosses can now be directed to prevent this from happening, and effects can be made to maximize the genetic variability in each generation. A potential genetic bottleneck has been recognized and can be avoided.
While the F1 fish can still be used in a genetic selection program, it will be important to include other sources of broodstock, such as the noncontributing parents, their offspring, or additional wild halibuts. Cooperation between hatcheries and the exchange of disease-free, pedigreed fish or gametes can also increase the genetic variability available for selective breeding programs. An effective population size of 13 is far too small to form the base of a selection program and could lead to potentially disastrous effects for the industry after only a few generations.
Genetic markers
In addition to their utility in monitoring genetic variation, micro-satellite markers allow one to determine pedigrees of fish whenever it’s convenient, provided tissue samples from the parents are available for comparison. Micro-satellites are also essential tools for the development of genetic maps and the identification of quantitative trait loci (QTL).
Genetic maps, which are constructed using hundreds of microsatellites, and QTL allow the identification of markers associated with economically important traits such as growth rate or disease resistance. Once identified, markers linked to QTL can be used to select fish with desirable traits at a very early age and thus increase the efficiency of the broodstock selection program.
(Editor’s Note: This article was originally published in the February 2004 print edition of the Global Aquaculture Advocate.)
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Authors
-
Michael E. Reith, Ph.D.
NRC Institute for Marine Biosciences
1411 Oxford Street
Halifax, Nova Scotia, Canada
B3H 3Z1 -
Timothy R. Jackson
NRC Industrial Research Assistance Program
University of New Brunswick
St. John, New Brunswick, Canada -
D. J. Martin-Robichaud
Fisheries and Oceans Canada
St. Andrews Biological Station
St. Andrews, New Brunswick, Canada
Tagged With
Related Posts

Intelligence
Halibut farming in Norway
While stable production of juveniles remains an issue for halibut farming in Norway, advances such as diets to replace live artemia are improving the industry.

Intelligence
Farmed white halibut
Farmed white halibut initially undergo an 18-month hatchery and nursery stage in land-based tanks. The fish have gained status as a high-quality seafood delicacy.

Health & Welfare
Green water culture of California halibut larvae
Although aquaculture offers potential for the production of California halibut, several aspects of production have yet to be optimized.

Innovation & Investment
Norway showcases halibut farm as sustainability star
Recognizing its potential as a sustainable fish producer, Norway highlights land-based halibut farmer Sogn Aqua in the debute of The Explorer digital showroom. The Advocate recently paid a visit to the farm in Ortnevik.


