A DNA sensor detected as low as 5 percent adulteration of European pilchard with round sardinella

Food fraud is a major problem worldwide, and because it is linked to economic interests and health concerns, food fraud is a high priority for food safety and quality worldwide. In addition, consumer demand for correct food labeling is constantly increasing. Along with other animal products, fishery products are considered to be one of the most adulterated foods. In addition, fish are among the most easily adulterated foods due to morphological changes during processing that are not visible to the naked eye.
Adulteration is the substitution of one species of fish with another species of lower price and lower quality. Species swaps comprise the most prevalent fish fraud. Sardines are vulnerable to adulteration as they contain valuable nutrients and are widely consumed worldwide. The only species that can be listed as sardine in canned food is the European pilchard (Sardina pilchardus), while if other similar species are used such as the round sardinella (Sardinella aurita), the trade name of “X sardines,” where X is different from S. pilchardus species, must be used.
To control adulteration, various analytical methods have been developed, including spectroscopy, chromatography, and protein-based methods. However, these methods are typically time-consuming, require highly trained personnel, and often use expensive and complex instrumentation and chemometrics for data interpretation. DNA-based methods are preferred for fish identification, especially for canned fish, because DNA is characteristic of each species and resistant to food processing conditions, such as the heat treatment during canning. Therefore, DNA-based methods are considered valuable analytical tools against food fraud.
This article – summarized from the original publication (Kakarelidou, M. et al. 2024. Fish DNA Sensors for Authenticity Assessment–Application to Sardine Species Identification. Molecules 2024, 29(3), 677) – reports on a study to develop fish DNA sensors in a simple rapid-test format, which enable visual identification of the two main sardine species, Sardina pilchardus and Sardinella aurita.
Study setup
In this study, a molecular rapid test using DNA sensors was developed for the detection of fish adulteration for the sardine species S. pilchardus with S. aurita. DNA was first isolated from the fresh tissue of both fish species, as well as from processed mixtures of the two species. The DNA was subjected to amplification by PCR using a common primer pair for the two species. The PCR products were analyzed by electrophoresis and quantification of the products was performed using the free-online ImageJ-gel Analyzer Software of the National Institutes of Health, and the DNA fragments were compared with a commercial DNA marker.
The sensing principle involves species recognition, via short hybridization of PCR-amplified sequences with specific probes, capture in the test zone of the sensor, and detection by the naked eye using gold nanoparticles as reporters; thus, avoiding the need for expensive instruments. In contrast to other methods, the proposed method provides shorter analysis time, and allows the detection of as low as 1 percent adulteration in mixtures of PCR products and 5 percent adulteration in tissue mixtures of processed (canned) samples of both species.
For detailed information on the experimental procedures, collection and analyses of samples, and analytical techniques used, refer to the original publication.
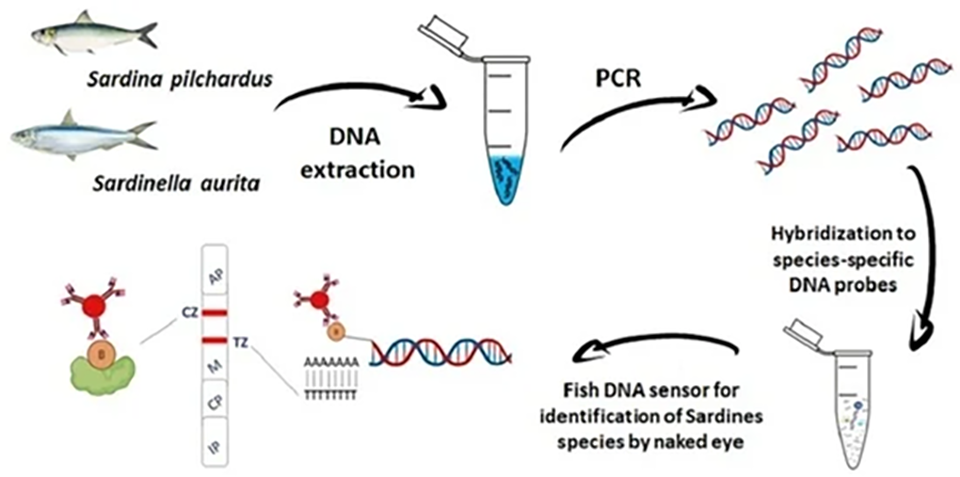
Results and discussion
In this study, a molecular rapid test was developed for the detection of fish adulteration for the species S. pilchardus with S. aurita. The identification of the two sardine species was performed using two different species-specific DNA probes that were hybridized to the amplified sequences followed by detection with the fish DNA sensor. The principle of the method is illustrated in Fig. 2.

Fish authentication is a major concern worldwide. For these reasons, new methods for fish species identification are of great importance. DNA-based methods are the preferred ones due to DNA stability and enhanced sensitivity. This study developed a DNA sensor for visual discrimination of S. pilchardus from each most common adulterant S. aurita. As low as 5 percent adulteration of S. pilchardus with S. aurita was detected with high reproducibility in the processed mixtures simulating canned fish products.
So far, few methods have been reported for authenticity testing of sardines. All these methods usually require expensive instrumentation or are highly time-consuming. The DNA sensor method reported here enables rapid and visual discrimination by the naked eye with high discrimination capability. However, it can only provide semi-quantitative results. The sensor is easy to develop and use, without the need for special and expensive instrumentation. Therefore, it can be easily adopted by any laboratory and public authorities for authenticity testing. Also, one of the great advantages of the proposed DNA sensor is its universality and versatility.

The sensor can be used for the identification of any other fish species as the molecular recognition by species-specific primers and probes is performed prior to the application to the sensor. Finally, the proposed DNA sensor can be modified accordingly to be able to detect more than two different species-specific DNA sequences on the same sensor.
Can handheld DNA testing technology stand up to seafood fraud?
Perspectives
The authors developed a fish DNA sensor for fish authentication in a rapid-test format. The method involves detecting adulteration of European pilchard with round sardinella using species-specific DNA probes. The developed DNA sensor is the first DNA sensor reported for the identification of sardine species and particularly for the detection of adulteration of S. pilchardus.
Detection is performed visually using gold nanoparticles as reporters. Compared to existing DNA-based fish adulteration methods, the method is simple and rapid. The sensor is cost-effective, easy to use, and very practical as it can be developed in the form of a dry-reagent strip-type sensor; no qualified personnel or costly instrumentation is required, while it can be used by authorities for fish authentication control or any laboratory including fish processing industries.
The method was successfully applied to fresh and processed samples treated by cooking and salting in the presence of other ingredients to simulate canning conditions. For mixtures of the two species, as low as 5 percent adulteration was detected in the processed samples with very good repeatability.
Now that you've reached the end of the article ...
… please consider supporting GSA’s mission to advance responsible seafood practices through education, advocacy and third-party assurances. The Advocate aims to document the evolution of responsible seafood practices and share the expansive knowledge of our vast network of contributors.
By becoming a Global Seafood Alliance member, you’re ensuring that all of the pre-competitive work we do through member benefits, resources and events can continue. Individual membership costs just $50 a year.
Not a GSA member? Join us.
Author
-
Theodore K. Christopoulos, Ph.D.
Corresponding author
Analytical/Bioanalytical Chemistry & Nanotechnology Group, Department of Chemistry,University of Patras, Rio, 26504 Patras, Greece; and 3 Institute of Chemical Engineering Sciences, Foundation for Research and Technology Hellas (FORTH/ICE-HT), Platani, 26504 Patras, Greece
Tagged With
Related Posts

Intelligence
‘Through science, there’s no question’: How evidence-based transparency is changing seafood traceability
ORIVO, a science-based testing and certification service for the global feed and supplement industry, aims to change seafood traceability.

Intelligence
What the COVID-19 pandemic taught us about seafood fraud
A co-author of a recent paper discusses two innovative case studies emerging from work conducted during the COVID-19 pandemic to minimize seafood fraud.

Intelligence
In the fight against seafood fraud, the technology behind trace element fingerprinting is maturing
Analysis of trace chemical elements can reveal where farmed seafood comes from. Standardization of traceability tools and techniques is improving the process.
Health & Welfare
Microsatellites: Genetic markers of choice
Use of microsatellites is one of many marker approaches, but has proven to be the tool of choice across all commercial agriculture sectors.



